The prenatal origins of cancer (original) (raw)
. Author manuscript; available in PMC: 2014 Oct 1.
Published in final edited form as: Nat Rev Cancer. 2014 Mar 6;14(4):277–289. doi: 10.1038/nrc3679
Abstract
The concept that some childhood malignancies arise from postnatally persistent embryonal remnant or rest cells has a long history. Recent research has strengthened the links between driver mutations, and, embryonal and early postnatal development. This evidence, coupled with much greater detail on the cell of origin and the initial steps in embryonal cancer initiation, has identified important therapeutic targets and provided renewed interest in strategies for the early detection and prevention of childhood cancer.
Introduction
Human cancer is a multistep process, evolving over many years inexorably toward genomic instability and life-threatening cellular phenotypes. The recognition that a minority of cancer cells within a tumour possess potent embryonal features, or the plasticity to revert to embryonal features during postnatal life, has generated the cancer stem cell hypothesis1, reflecting a tumour’s almost limitless heterogeneity. Cancer stem cells are not replicates of their embryonal counterparts, but instead have some features of embryonal cells that provide a survival advantage suited to a particular time and place during tumour evolution1. By contrast, cancers presenting very early in a child’s life often possesses embryonal features but must have moved quickly to genomic instability. This rapid evolution might occur when a cancer cell arises directly from an embryonal cell, or when a mature prenatal cell acquires embryonal properties which favour survival in the pre- and postnatal environment. Prenatal tumorigenic mutations might be more likely to occur in stem or progenitor-like cells that have characteristics, such as unrestrained self-renewal, that are essential in utero but fatal in postnatal life. Understanding the characteristics of these cells might well be important to understanding more about childhood cancer: why it develops, how best to treat it, and potentially how to prevent it.
For some childhood cancers, such as retinoblastoma, infant acute lymphoblastic leukaemia (ALL) and malignant rhabdoid tumours, there is evidence of an embryonal cell of origin. However, it is unclear whether these embryonal cancers arise from embryonal cells in utero or as a consequence of a single oncogenic event in a more mature prenatal cell with rapid progression to genomic instability. The latter model is likely to be the case for mixed lineage leukaemia (MLL) fusion genes in infant ALL, and INI1 mutations in malignant rhabdoid tumours9, 10.
A unique characteristic of certain childhood cancers, such as neuroblastoma and Wilms tumour, is a precursor embryonal cell hyperplasia that presents in infancy but that has an overwhelming tendency to undergo spontaneous regression and cell death4, 5. More than 150 years ago pathologists first suggested that postnatally persistent embryonal remnant or ‘rest cells’ lay dormant in normal tissues but retained their capacity for growth and might later undergo malignant transformation6–8. Today, we know that during embryogenesis many more cells are produced than are required for organogenesis. Thus mechanisms, such as trophic factor withdrawal, are required for deleting cells in excess once organogenesis is complete. In rare instances, some embryonal cells can resist these cell death signals as a potential first pathological step toward cancer, later acquiring additional alterations which lead to postnatal malignant transformation.
Some types of cancer with embryonal histology, such as germ cell tumours, more commonly arise in early adulthood, however, it is unclear whether these malignancies represent cellular reversion from a mature to an embryonic form in adult life or postnatal persistence of embryonal cells, since there is no evidence of a rest disease pre-dating these cancers. Therefore, although the concept of rest cells is helpful in considering the developmental processes that might underlie cancer development in young children, lack of evidence for a rest disease before malignant transformation has been taken as evidence that few childhood cancers arise through this mechanism. However, a substantial number of childhood malignancies seem to arise prenatally. If all childhood malignancies with a suspected prenatal cell of origin are grouped together it is apparent that almost half of all childhood cancers might have prenatal origins (Table 1). In the case of B-lineage ALL (B-ALL), transient myeloproliferative disorder (TMD) and myeloid leukaemia-Down syndrome (ML-DS), driver mutations and causal gene rearrangements or balanced translocations are detectable in perinatal peripheral blood leukocytes years before clinical presentation, indicating these childhood cancers initiate in utero2, 3.
Table 1.
Embryonal childhood malignancies
| Malignancy | Tissue of Origin | Evidence for prenatal origin | |
|---|---|---|---|
| Pre-Clinical | Clinical | ||
| Neuroblastoma | Neural crest sympathoadrenal progenitors (neuroblasts) | -Transgenic expression of genes involved in sympathoadrenal development (MYCN, ALK, LIN28B) recapitulate neuroblastoma20, 33, 43. -Animal modeling with Th-MYCN mice demonstrate rest formation prior to tumour formation due to resistance to developmental deletion signals15, 19. -Neural crest stem cell allografts generate neuroblastoma with oncogene (MYCN, ALK F1174L) transduction44. | -Young age of incidence (<5 years)12. -Multifocal/bilateral primary tumours in tissues derived from the embryonic neural crest in some 20% patients11. -Normal foetal neuroblasts highly similar to tumour derived neuroblasts45. -Failure of complete sympathoadrenal maturation - Neuroblast rests and elevated catecholamines (neuroblastoma marker) in subclinical patients46, 47, 172. -Low stage neuroblastoma and rest cells frequently regress4, 46. |
| Wilms Tumour | Primitive metanephrogenicblastema | -Young median age of incidence173. -Wilmstumour can be bilateral173, 174. -Inactivating mutations in kidney development gene WT1 predispose for Wilm’stumour173. -Nephrogenic rests adjacent to tumours174. -WT-1+ nephrogenic rests found adjacent to WT-1+ tumours174. -Nephrogenicrests can spontaneously regress5, 175. | |
| Medulloblastoma | Neural progenitors of the developing cerebellum/brainstem | -Aberrant SHH signaling through development recapitulates SHH subgroup medulloblastoma114, 121, 122. -SHH medulloblastoma modeling exhibit pre-malignant rests prior to tumour formation due to resistance to developmental cell deletion signals117. -Activating mutations in β-catenin and aberrant WNT signaling in OLIG3+ neural precursors through brainstem development can recapitulate medulloblastoma137. -Animal modeling shows that cerebellar development genes (MYCN, MYC) can recapitulate medulloblastoma in SHH-independent medulloblastoma126–128. | -SHH subtype associated with developmental disorder Gorlin Syndrome111. -Allelic loss on chromosome 9 (containing SHH pathway suppressor PTCH1) gene in 30% of tumours113. |
| Retinoblastoma | Retinal progenitors of the developing eye | -Inactivation of retinal development genes Rb1 and p107 in retinal progenitors causes prolonged proliferation of progenitors through development and is sufficient to recapitulate retinoblastoma176, 177. | -Familial cases are often bilateral, multifocal and usually occur in the first 2 years of postnatal life143, 178. -Retinal progenitor cell proliferation occurs only in foetal retina178. -Premature baby reported with retinoblastoma in early postnatal life179. -Patients are defined by familial and sporadic inactivation of retinal development gene Rb1180. |
| Germ Cell Tumour (GCTs) | Primordial germ cells (PGGs) originate in yolk sac endoderm | -Subset of patients with early incidence < 4 years181, 182. -Teratomas can be found prenatally183. -Germline genetic predisposition have earlier onset181. -Bilateral GCTs181. -Genetic aberrations such as loss of 1p and gain of 1q are found in paediatric GCT which may reflect loss of tumour suppressor gene on 1p, involved in terminal differentiation of embryonic tissues182. -Malignant GCT component found within ‘benign’ teratomacomponent, reflects stages of malignant transformation182. -Carcinoma-in-situ cells resemble primordial germ cells in adult onset testicular GCT’s181. | |
| B-ALL | Haematopoietic system (Lymphoid-primed multipotent progenitors LMPPs) | -Pre-leukaemic population detected in utero in TEL-AML transgenic mice86, 87, 184. | -Twin studies of monozygotic twins with concordant leukaemia (TEL-AML1 positive pre-B ALL, infant ALL)79, 81. -Twin studies: clonal evolution of in utero pre-leukaemic population in neonatal blood spots in TEL-AML181, 86. -Pre-leukaemic cells detected in neonatal blood spots for majority of pre-B ALL patients2, 82. |
| TMD and ML-DS | Haematopoietic system (Megakaryocyte-Erythroid Progenitors MEPs) | -GATA-1s knock-in mouse model of TMD demonstrate transient hyperproliferation in megakaryocytic progenitors in embryonic yolk sac and foetal liver67. | -Tissue of aborted 2nd trimester foetuses with Down syndrome demonstrating pathological increase in MEPs in foetal liver, prior to acquisition of somatic GATA1 mutation54. -GATA1 mutation detected in utero66. -In utero TMD diagnosis; condition present in preterm neonates53. -Median age of diagnosis 3–7 days postnatally, range 0–65 days52, 53. -TMD regresses prior to progression to ML-DS in a 25% of patients49. |
| Malignant Rhabdoid tumours (MRTs) | Unknown cell of origin. Frequently arises in the kidney and central nervous system | -Heterozygous mutations of embryonic development regulator INI-1 in transgenic mice cause MRT predisposition and upon loss of heterozygosity manifests as MRTs185. | -Fetal and neonatal MRTs frequently identified183. -Concomitant primary tumoursin both the kidney and brain183. -Familial or sporadic inactivation of INI-1 gene apparent in >95% of tumours10, 186–188. |
Recent studies using genetically altered animal models, family linkage analyses, large scale expression profiling and genome-wide association studies (GWAS), have provided a rich vein of genes as possible drivers in prenatal cancers, many of which also have a role in embryonal development. This Perspective article focuses on four childhood malignancies with, we argue, the strongest evidence for a prenatal cell of origin: neuroblastoma, TMD and ML-DS, B-ALL and medulloblastoma. We discuss their prenatal origins in the context of normal tissue development and the potential cell of origin, as well as in the context of rest disease. Although rest cells are often seen as normal, we propose broadening the definition of rest disease to include any pre-malignant cellular hyperplasia detectable in the perinatal period. In this new definition, rest cells are already on the path towards malignancy, but these cells retain the compulsion to die in the postnatal environment, as they still require additional postnatal changes for malignant transformation. The aim of suggesting this new definition is to facilitate discussion and experimentation aimed at identifying common aetiological features, strategies for early diagnosis and therapy, and, possibly prevention of childhood cancers of prenatal origin.
Neuroblastoma
Neuroblastoma is a malignancy of the sympathetic nervous system occurring almost exclusively in infancy and early childhood11, 12. Primary neuroblastoma arises in the adrenal medulla or sympathetic ganglia along the paravertebral axis. Considerable evidence suggests that neuroblastoma is initiated in utero during sympathoadrenal development and is a rest disease.
Candidate neuroblastoma initiation factors linked to sympathoadrenal development
The cells of the mature adrenal medulla and sympathetic ganglia originate in a transient collection of neural crest progenitors that rely on spatially and temporally regulated extrinsic signals for the later steps of migration, specification, divergence and maturation13 (Figure 1). Sympathoadrenal cells from the neural crest in the trunk region of the embryo follow a ventral migratory pathway from the neural crest and neural tube, receiving signals from somites, ventral neural tube, notochord and dorsal aorta13. During normal sympathoadrenal development, expression of the proto-oncogene MYCN is high in the early post-migratory neural crest where it regulates the ventral migration and expansion of neural crest cells14. MYCN protein levels gradually reduce in differentiating sympathetic neurons14, 15, 16 suggesting that sympathoadrenal maturation requires low or absent MYCN expression14. Consistent with this finding is the observation that MYCN transduction into quiescent rat sympathetic neurons reactivates cell cycling and blocks cell death induced by nerve growth factor (NGF) withdrawal15,16.
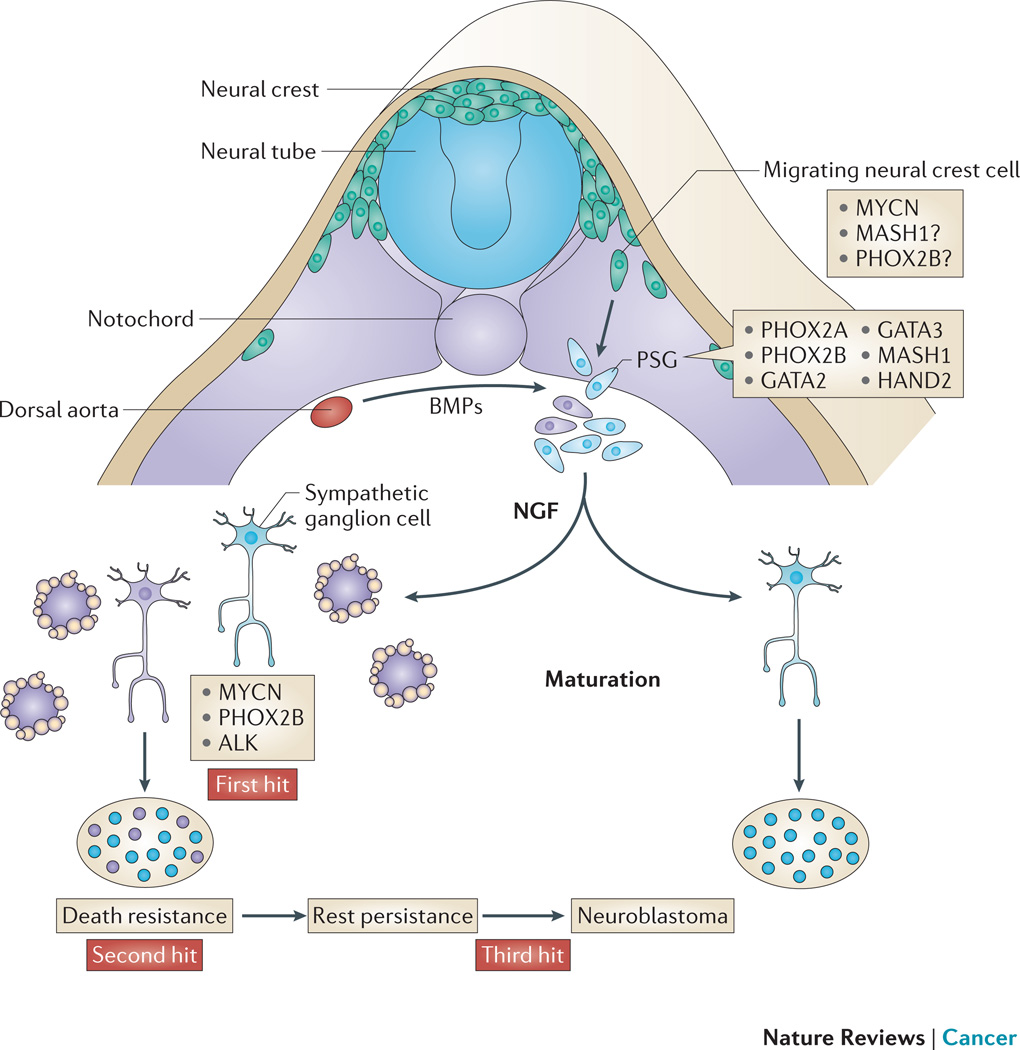
Figure 1. Neural crest development and neuroblastoma.
Neuroblast progenitors migrate from the neural crest (nc), around the neural tube (nt) and somites (so) to a region immediately lateral to the notochord (no) and the dorsal aorta (da) under the influence of MYCN and bone morphogenetic proteins (BMPs). At this site the cells undergo specification as the primary sympathetic ganglia (psg) before divergence into neural cells of the mature sympathetic ganglia (sg) or chromaffin cells of the adrenal medulla (am). MYCN is a first hit by virtue of the observations from the tyrosine hydroxylase (Th)-MYCN transgenic mouse model, whereas mutations/alterations in anaplastic lymphoma kinase (ALK) and paired-like homeobox 2 (PHOX2B) are inherited human susceptibility genes. Local access to nerve growth factor (NGF) determines whether the normal sg matures into a terminal ganglion cell or undergoes apoptotic cell death. A relatively common pathologic state is postnatal survival of neuroblast rest disease which requires the cell destined to become malignant to be resistant to trophic factor withdrawal before these persistent rest cells undergo a third change to induce transformation, which presents as a clinical malignancy in early childhood. MASH1, murine achaete-scute homolog 1; HAND2, heart and neural crest derivatives expressed 2; NF-M, neurofilament medium polypeptide.
After sympathoadrenal specification and expansion of the primary sympathetic ganglia, sympathoadrenal precursor development diverges into neuronal or chromaffin cell fates13. Based on expression patterns of differentiation markers, the earliest cell of origin for neuroblastoma is thought to be a neural crest cell specified to the sympathoadrenal lineage, which has not received or responded to cues that determine neuronal or chromaffin cell fate (Figure 1). Excess neural precursors undergo apoptotic cell death at the final stage of sympathoadrenal maturation17, a process catalysed by local NGF deprivation. In zebrafish, persistent MYCN expression in sympathoadrenal precursor cells dramatically blocks development toward a chromaffin cell fate, leading to neuroblastoma18. Overexpression of human MYCN under the control of the rat tyrosine hydroxylase (Th) promoter in Th-MYCN transgenic mice is sufficient to recapitulate neuroblastoma rest disease (discussed in humans below) and tumorigenesis15, 19, 20. Transformation from a rest cell to a malignant neuroblast in Th-MYCN mice requires even higher levels of MYCN than are provided by the transgene. This occurs via transgene amplification and feed forward loops between MYCN and increased levels of NAD-dependent deacetylases, sirtuins 1 and 2, which increase MYCN stability21, 22.
This cellular tolerance for high-level MYCN is surprising as supra-physiological levels of MYC expression should lead to apoptosis and senescence through the cyclin-dependent kinase inhibitor 2A, (CDKN2A) isoform 4 (known as ARF)–p53 stress response pathway23. Rest cell cultures from perinatal Th-MYCN mouse ganglia demonstrate lower basal and induced p53 levels than wild-type ganglia, which in turn are lower than p53 levels of mature ganglia19. When p53 is reactivated in rest cell cultures, the cells become sensitive to NGF deprivation19. Inactivation of the ARF–p53 pathway in neuroblasts can be accomplished by the Polycomb complex protein BMI1, acting as a p53 protein E3 ubiquitin ligase in Th-MYCN mice, or mutant anaplastic lymphoma kinase (ALK), through effects on PI3K and MAPK signalling in zebrafish19, 24. These findings indicate that there might be an inherent susceptibility to oncogenic stress in some embryonal cells lacking robust p53 stress responses.
These experiments mechanistically connect MYCN with the maintenance of neuroblast rests and as a driver of resistance to developmentally-timed trophic factor withdrawal signals at neuroblastoma initiation. The marked downregulation of high affinity nerve growth factor receptor (NTRKA) in MYCN amplified neuroblastoma cells blocks neural differentiation. Conversely, NTRKB responds to brain-derived neurotrophic factor (BDNF) which has survival- and growth-promoting actions on neurons. BDNF and NTRKB are expressed by MYCN-expressing neuroblastoma cells and maintain an autocrine cell survival loop that blocks differentiation and apoptosis11.
Five weeks into human development, the ventrally migrating neural crest cell responds to bone morphogenetic protein signalling from the dorsal aorta to promote a transcriptional program that specifies the neuronal and catecholaminergic properties of the expanding primary sympathetic ganglia13, 25, 26. Murine achaete-scute homologue 1 (MASH1) then promotes paired-like homeobox 2A (PHOX2A) expression which, together with PHOX2B, drives the expression of enzymes for catecholamine biosynthesis. PHOX2B initiates a secondary transcriptional program, which controls terminal sympathoadrenal differentiation. Heterozygous germline mutations of PHOX2B are associated with a subset of familial neuroblastoma27, 28. Neuroblastoma driven by PHOX2B mutations often presents clinically in conjunction with the neural crest disorders Hirschsprung’s disease and congenital hypoventilation syndrome, reinforcing the connection between neuroblastoma and defective neural crest development. PHOX2B mutant proteins appear to be oncogenic in a dominant negative manner and promote inappropriate proliferation in neural precursors29, 30. Interestingly, rest cells from Th-MYCN mice have high PHOX2B expression, indicating a possible indirect downstream link to MYCN31. Future experiments will uncover the finer detail of the regulatory relationships between these proteins in development and disease.
Paralogous RNA-binding proteins LIN28A and LIN28B regulate expression of the let-7 miRNA family during embryogenesis, controlling developmental timing and cell growth during neural crest cell lineage commitment32. Targeted expression of LIN28B to the developing neural crest of transgenic mice induces neuroblastoma33. LIN28B downregulates let-7 pre-miRNA’s, which increases MYCN expression33. LIN28B is known to be crucial in the maintenance of stemness throughout embryonic development and this function might maintain the undifferentiated neuroblast phenotype33, 34. It will be important to determine whether LIN28B transgenic mice recapitulate the rest disease seen in Th-MYCN mice. The known role of LIN28B in neurogenesis35 fits well with a model of neuroblastoma initiation requiring aberrant regulation of developmental proteins. LIN28B can be amplified and highly expressed in some neuroblastoma tumours, which correlates with poor prognosis33. Interestingly, LIN28B–let 7 signalling through MYCN seems to be important in another putatively embryonal cancer, germ cell tumour36.
ALK has a key role in early sympathoadrenal development to protect neuroblast growth in utero against nutrient deprivation37, 38. Germline and somatic mutations in ALK causing constitutive kinase activation are present in 8–10% of neuroblastoma cases and correlate with poor prognosis39–42. The most common and aggressive activating mutation of ALK, F1174L, is sufficient for tumour formation on neural crest-specific expression in transgenic mice43 and on expression in neural crest cells transplanted into nude mice44. ALK-F1174L has been associated with MYCN amplification in human tumours and co-expression of ALK-F1174L and MYCN synergistically promotes tumour formation in vivo suggesting these may be co-operating events in tumour initiation43. Thus, aberrant MYCN expression or function in collaboration with disordered function of a small group of key neurodevelopmental regulators, are good candidates for neuroblastoma rest disease initiation. The exact order of these events, or the intrauterine factors which favour rest disease, are unknown.
Neuroblastoma as a sympathoadrenal rest disease
Several clinical and experimental features link neuroblastoma to defective embryogenesis and pathological rest disease. The embryonal origin of human neuroblastoma is supported by expression profiling showing that human foetal adrenal neuroblasts have gene signatures that are remarkably similar to neuroblastomas45. In some neuroblastoma patients new tumours form at different times and sites early in the child’s life, suggesting a “field” of pre-malignant lesions, thus linking tumorigenesis to deregulated development.
A key feature of clinical or experimental rest disease is the compulsion to undergo cell death and spontaneous regression in the postnatal environment, thus mirroring features of embryonal cells in utero not required for organogenesis. The incidence of subclinical neuroblastoma and/or rests is much higher than the frequency of clinical disease. A small proportion of neuroblastoma patients present in early infancy with apparently metastatic tumour (stage 4S neuroblastoma) which undergoes regression without therapy. Autopsies of sympathoadrenal tissues from infants who have died of diagnoses other than cancer have shown an incidence of neuroblast rests 40-fold higher than the clinical disease46. Mass infant screening programs in the 1990s for the neuroblastoma tumour marker, urinary catecholamines, found subclinical tumours at a frequency more than 2-fold higher than the clinical incidence of neuroblastoma47.
These mass screening programs aimed to prevent the later incidence of metastatic neuroblastoma by early tumour detection in infancy at the localised stage, followed by surgery, but were unsuccessful in this regard47. One explanation is that rest cells which later progress to metastatic neuroblastoma might already be fundamentally different at birth from the more common spontaneously regressing rest disease. This hypothesis is suggested by work in Th-MYCN transgenic mice, showing a small number of perinatal rest cells, later selected for tumour formation, had undergone early MYCN transgene amplification15. Moreover, recent transgenic zebrafish modelling of neuroblastoma also indicates that there is a need for a co-operating signal, inactivating MYCN-induced apoptosis at tumour initiation18. Animal modelling confirms metastatic neuroblastoma can arise from rest cells15.
The postnatal period of quiescent neuroblast rests in sympathoadrenal tissues offers a time window during early infancy when alternative forms of screening for rest disease might be envisaged based on a germline cancer susceptibility profile and recently described methods of detecting tumour DNA in peripheral blood48. Furthermore, it might be expected that drugs which restore the p53 stress response in rest cells, given briefly at this crucial early tumour phase might be effective in restoring the capacity for spontaneous regression and thus preventing malignant transformation.
Another prenatal cancer and potential rest-like disease that offers significant opportunities for early detection and therapy is TMD and ML-DS.
TMD and ML-DS
TMD is a megakaryocyte (MK) lineage disease that presents clinically at birth in 5–10% of children with DS, and has similarities to embryonal rest disease states49, 50. The clinical abnormalities associated with TMD resolve spontaneously in early infancy in almost all affected patients, however, approximately 20% of these children present 2–4 years later with ML-DS, an acute MK lineage leukaemia49, 51.
MK development and TMD pathogenesis
TMD develops in utero during megakaryopoiesis. Megakaryocyte progenitors (MKPs) are first produced from haematopoietic stem cells (HSCs) in the yolk sac, aorto-gonad-mesonephros and thereafter the foetal liver52–54. Rarely, TMD can occur in infants without DS but all of these cases have trisomy 21 mosaicism in the marrow, supporting the key role of trisomy 21 in TMD pathogenesis. Moreover, the frequency of conversion to ML-DS is the same for patients with TMD linked to DS or non-DS trisomy 21 mosaicism50, 52, 53, 55. Foetal liver haematopoiesis in DS shows specific expansion of HSC and MKP compartments57,58. These trisomy 21-induced changes are likely to occur during definitive haematopoiesis from the yolk sac or aorto-gonad-mesonephros57, 58. Ts65Dn mice carry an extra chromosome with gene copies of most of mouse chromosome 16, which is orthologous to human chromosome 2156. Ts65Dn mice do not develop TMD spontaneously, but display abnormal megakaryopoiesis and chronic myelofibrosis56.
MK development and differentiation is controlled by combinations of cytokines and other mediators present within a haematopoietic vascular and endosteal niche58, 59. Of the genes on chromosome 21, several are good candidates for leukaemogenesis: runt-related transcription factor 1 (RUNX1)ERG, ETS2, dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (DYRK1 Α) and GA binding protein transcription factor-α (GABPA). RUNX1 expression is not increased in ML-DS60 and trisomy of Runx1 was not required for the development of a myeloproliferative disorder in Ts65Dn mice56. The ETS transcription factor ERG, on the other hand, can immortalise haematopoietic progenitors61, 62, and cooperate with a truncated version of GATA1 to generate a TMD-like defect in vivo63.
RUNX1 interacts with additional factors, including the transcriptional activator GATA164. GATA1 levels increase and GATA2 levels decrease during MK differentiation59. The GATA1 gene is mutated in megakaryoblasts in all cases of TMD and ML-DS and these mutations, in exon 2 or at the exon 2–intron boundary of GATA1, lead to exclusive production of truncated GATA1, termed GATA1s65 as early as 21 weeks of gestation66. GATA1s does not have an N-terminal transactivation domain, but retains both DNA-binding zinc fingers, and its expression leads to impairment of GATA1-mediated regulation of other transcription factors including GATA2, IKAROS family zinc finger 1 (IKZF1), MYB and MYC in foetal MKs67. Paired sample analysis by several groups indicates that the same GATA1 mutation is present in TMD and matched ML-DS samples53, 68–70. However, neither the mutation site nor the nature of the mutation in GATA1 exon 2 predicts progression to ML-DS70 and surprisingly, Kanezaki et al found an association between low levels of GATA1s protein and a higher probability of progression to ML-DS71.
TMD as a rest disease
TMD develops in utero when blood cell production resides in the foetal liver52, 53, 72, 73. Trisomy 21 creates MKP hyperplasia, and increased megakaryocyte early progenitor (MEP) clonogenicity, which precedes GATA1s expression54, 57, 74. Elegant mouse studies of the effects of a GATA1s knock-in allele showed that GATA1s expression leads to transient hyperproliferation of an embryonic MKP beginning in the yolk sac, then moving to the foetal liver, ultimately disappearing once haematopoiesis moves to the bone marrow postnatally67. Foetal liver-derived megakaryoblasts have been recently shown to differ from their adult counterparts by their low type 1 interferon production: type 1 interferons are known inhibitors of MKPs. This might help to explain why trisomy 21 and GATA1s have specific effects on foetal MKPs75, and also indicates that exogenous interferon α might be an effective deletion therapy for residual foetal MKPs in TMD.
Thus, to date the evidence indicates that the progression from normal prenatal MKP to TMD requires trisomy 21 as the ‘first hit’49 and then somatic GATA1 mutation as the ‘second hit’. We think that it is likely that trisomy 21 and GATA1s combine to promote a perinatal TMD rest-like disease in some DS children by causing inappropriate MKP hyperplasia in the foetal liver. Like other rest diseases, MKP rest-like cells retain the compulsion to die in the perinatal period by as yet unknown mechanisms once haematopoiesis shifts to the bone marrow. At birth the TMD rest-like cells need a ‘third hit’ to sustain viability in the face of death signals. Whether these death signals also involve p53 is unknown. Transcription profiling studies of TMD and ML-DS samples indicate that driver mutations which activate WNT, JAK-STAT and MAPK–PI3K signalling are good candidates for a ‘third hit’76 (Figure 2). Furthermore, recent TMD patient xenograft analyses demonstrate distinct subclones emerging on serial transplantation with genomic features of ML-DS, such as chromosome 16q deletion and chromosome 1q gain, which were present at low frequency in the initial TMD sample77. These observations are reminiscent of the process of clonal selection for transgene amplification seen in Th-MYCN transgenic mouse neuroblast rests15, and support the idea that even at birth rare leukaemogeneic or tumorigenic subclones exist among apparently regressing rest-like cell populations.
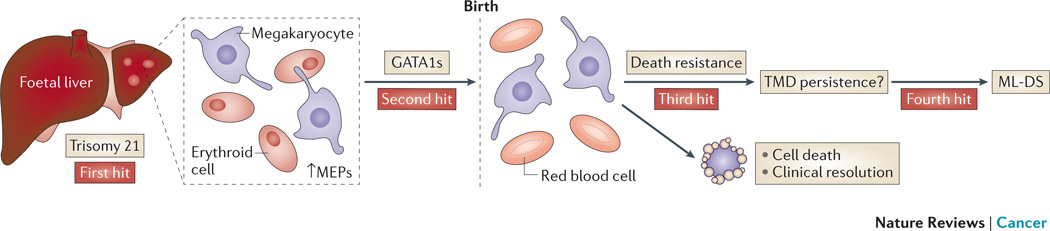
Figure 2. TMD and ML-DS.
Children with Down Syndrome (DS) can develop Transient Myeloproliferative Disease (TMD) at birth which can later transform to Myeloid Leukaemia (ML-DS). The production of megakaryocyte-erythroid progenitors (MEPs) is enhanced in the foetal liver of children with DS and trisomy 21 as a first hit. Some megakaryocyte precursor cells develop a second hit mutation in GATA1 resulting in a mutant, truncated protein, GATA1s. All children with TMD and ML-DS exhibit GATA1s and more than two copies of chromosome 21. TMD resolves clinically in almost all patients to later present as ML-DS in 25% of TMD cases. Thus, some TMD cells at birth must survive through unique death resistance mechanisms and may undergo further changes which lead to genomic instability and clinical presentation as ML-DS.
Collectively, we think that these data suggest that rest cells are not normal embryonal cells that have simply persisted postnatally. Instead postnatal rests, like cancer stem cells, have retained embryonal features at the early stages of a traditional pathway towards tumorigenesis. Recent studies using next generation sequencing of peripheral blood DNA from DS neonates to analyse for GATA1 mutations indicates the incidence of TMD may be in the range of 30%78. These findings suggest a strategy of predictive testing for TMD in DS neonates and pave the way for therapeutic intervention studies for TMD rest-like disease as a method of leukaemia prevention.
Another childhood cancer where leukaemia marker genes have been identified in the perinatal period suggesting prenatal origins is B-ALL. However, the absence of a clinical B-ALL rest disease and the rarity of the premalignant clone in the perinatal period means that application of preventative disease strategies for B-ALL will be more difficult.
B-ALL
ALL is the most common childhood malignancy, and arises in B- or T-lineage lymphocyte progenitor cells. We confine our discussion to the more common B-lineage ALL and B cell development, since T-ALL generally presents in adolescence, and most of the evidence for a prenatal origin has been obtained using samples from children with B-ALL.
Leukaemogenesis begins prenatally in children with B-ALL
Leukaemic cells from identical twin pairs with concordant B-ALL share unique, clonal, non-constitutive chromosome rearrangements, even when diagnosed years apart, indicating that leukaemogenic molecular alterations occur in utero79, 80. Further support for the idea of a prenatal origin came from a study of 5-year-old monozygotic twins aimed at identifying leukaemogenic markers81. These patients presented with B-ALL and concordant translocation of ETS variant 6 and RUNX1 genes (also known as TEL-AML1). The leukaemic cells also showed distinct immunoglobulin heavy-chain (IgH) and immunoglobulin kappa deleting element (IgK-Kde) gene rearrangements in neonatal blood spot DNA, indicating that separate preleukaemic clones must have evolved before birth81. Using IgH rearrangement as a marker for leukaemic clones, several groups have now shown that pre-leukaemic cells can be detected in the neonatal blood spot of the majority of childhood B-ALL patients, indicating that the leukaemogenic process begins in utero2, 82 (Figure 3).
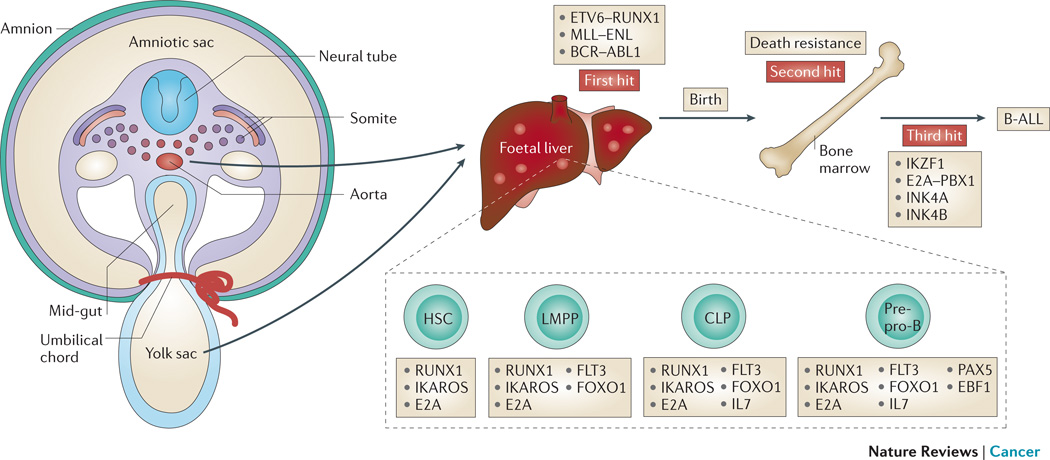
Figure 3. The development of B-ALL.
The presence of specific prenatal oncogenic fusion proteins in haematopoietic stem cells (HSCs) or B progenitor cells combines with aberrant expression of transcription factors necessary for normal B cell development and maturation during the genesis of acute lymphoblastic leukaemia (B-ALL). Foetal haematopoiesis begins in the haemogenic endothelium formed from the yolk sac and aorto-gonadal-mesonephros (AGM), then localizes to the foetal liver, to finally reside in the bone marrow from the perinatal period. Several fusion genes (ETV6-RUNX1, MLL-ENL, BCR-ABL) present as clonal chromosome rearrangements at the point of diagnosis of B-ALL, represent the first hit as they have also been identified in blood samples at birth in children who later develop B-ALL. Since the incidence of the ETV6-RUNX1 fusion gene is higher than the clinical incidence of B-ALL, some B-ALL rest cells must survive the perinatal period to undergo a third hit that leads to clinical presentation.
The TEL-AML1 fusion gene is a clonal feature of B-ALL cells at diagnosis in more than 20% of patients, but is present on neonatal blood spot analysis at a much higher frequency than the incidence of B-ALL in childhood83–85. Studies of a monochorionic _TEL-AML1_-positive twin pair (one preleukaemic and one leukaemic), have established that the presence of TEL-AML1 in B cells is sufficient to generate a population of pre-leukaemic cells and act as a ‘first hit’ mutation that results in altered self-renewal and survival properties86. However, low level TEL-AML1 expression only is tolerated by pro-B cells since these cells terminally differentiate in the long term87. Although TEL-AML1 has been found in peripheral blood leukocyte DNA from healthy adults (0.5–8.8%), it is much more frequent in young adults83, presumably representing pre-leukaemic clones which originated prenatally. TEL-AML1 expression is, of course, not part of the normal B cell developmental program. Thus, TEL-AML1 operates as a weak oncogenic event in prenatal HSCs or precursor B-cells to generate a pre-leukaemic state which, like precancerous neuroblasts and megakaryoblasts, have a short lifespan in the postnatal environment. TEL-AML1 may function as an oncogene only in neonates owing to specific characteristics of foetal B cells (Box 1), thus linking TEL-AML1-initiated leukaemia to the embryonal environment. Overexpression of the erythropoietin receptor, early B-cell factor 1 (EBF1), and deregulated transforming growth factor β responses, might have a role in survival of _TEL-AML1_-initiated pre-leukaemic cells88–90. Both the breakpoint cluster region (BCR)-ABL1 and mixed lineage leukaemia (MLL)-eleven nineteen leukaemia (ENL) fusion genes can occur prenatally and might initiate B-ALL91, 92. However, the pre-leukaemic clone also remains clinically silent in the absence of additional changes, such as an IKZF1 mutation91.
Box 1 Regulation of B-cell development and B-ALL leukaemogenesis.
Haemogenic endothelium from the mouse E9 yolk sac and the para-aortic splanchno-pleura independently give rise to the first B progenitor cells that preferentially differentiate into innate B-1 cells and marginal zone B cells, but not into adaptive B-2 cells150. Thereafter, B cells are generated from haematopoietic stem cells (HSCs) in the foetal liver until shifting to the bone marrow from birth onwards. Foetal liver-derived B cells exhibit some descriptive and functional differences from bone marrow-derived B cells, such as low terminal deoxynucleotidyl transferase expression, and a restricted diversity of the IgH variable and hinge region repertoire151. HSCs isolated from foetal liver can differentiate into B-1 and B-2 cells, however, B-1 production predominates until the bone marrow takes over and the B-2 postnatal program becomes established152. In embryonal or adult haematopoietic tissues, HSCs differentiate into lymphoid-primed multipotent progenitors (LMPPs) which express high levels of fms-related tyrosine kinase 3 (FLT3), lose megakaryocyte and erythrocyte progenitor (MEP) potential, and retain the capacity to differentiate into lymphoid and myeloid cells153. LMPPs can differentiate into early lymphoid progenitors, the precursors of bone marrow-derived common lymphoid progenitors (CLPs), which give rise to pre-pro-B cells expressing B cell lineage-associated genes154, 155. Mice deficient in FLT3 ligand have reduced CLP levels, whereas mice deficient in Flt3 and interleukin-7 receptor (Il7r) do not develop B cells156, 157.
A plethora of studies has revealed structural rearrangements, deletions, amplifications and point mutations in genes encoding regulators of B lymphocyte development in B-ALL patient samples, many of which are candidates for leukaemia initiation. Many are transcription factors that regulate B-cell development158. _IKZF1_-deficient HSCs exhibit reduced expression of IL-7R and FLT3 and loss of LMPPs, while reduced expression of IKZF1 in mice results in impaired transition from the pro-B cells to pre-B cells. Ikzf1−/− mice completely lack B cells159–161. IKZF1 is deleted in most BCR-ABL1-positive B-ALL cells and mice expressing mutant IKZF1 develop ALL162.
Conditional deletion of Pax5 in mature B cells leads to de-differentiation toward uncommitted HSCs163. Somatic deletion, mutation or rearrangement of PAX5 is detectable in more than 30% of children with B-ALL164. PAX5 DNA-binding and internal deletion mutants act as hypomorphic alleles with weak competitive activity164. Hypomorphic PAX5 mutations have recently been described in studies of two affected families with inherited B-ALL, thus representing familial B-ALL predisposition genes165.
E2A, which encodes the E protein transcriptional regulators E12 and E47, activates expression of forkhead box O1 (FOXO1), and is required for CLP formation and B lineage specification166–168. EBF1 is necessary for pre-pro-B cells to transit to the pro-B stage169. Forced overexpression of EBF1 in both wild type and _E2A_-deficient HSCs leads to B cell differentiation170.
RUNX1 is indispensable for generating CLPs171, and, the expression of PAX5, E12, E47 and EBF1 is reduced in B cell precursors lacking RUNX1170. Runx1 deficiency in mice leads to a severe reduction in CLPs, and a myeloproliferative phenotype171.
We hypothesise that B-ALL may also follow a traditional tumorigenic pathway which begins in prenatal, partially transformed cells which have adopted characteristics favoured by the embryonal, but not the postnatal, environment. Consistent with our proposed broader definition of rest diseases, B-ALL may also be preceded by clonal hyperplasia of a small transient B cell population which, like neuroblast rests and TMD rest-like cells are destined to die unless further changes accelerate the path toward malignancy. More detailed analysis of genetically modified animal models of B-ALL for rest hyperplasia, and its spontaneous regression, would strengthen the hypothesis that B-ALL begins as a rest disease. In this regard, the recent description of mutant p53 as another familial B-ALL predisposition gene suggests the p53 stress response may be defective in B-ALL rests, much like neuroblasts from the Th-MYCN mice15, 19, 93, 94. However, for B-ALL to be considered as a rest disease, several important facts need to be established: the exact embryonal timing and tissue of initiation, and the leukaemogenic order and relative inter-dependency of the candidate initiator mutations with B cell homeostasis in the prenatal environment. The susceptibility of foetal B cells to the weak TEL-AML1 oncoprotein might be due to the different cell intrinsic responses of HSCs or innate B-1 precursor cells, or cell extrinsic susceptibilities of the foetal liver compared with the postnatal bone marrow (Box 1).
More evidence for rest-like disease in another embryonal cancer, medulloblastoma, exists in mouse models. However, like B-ALL there is no clinical presentation of a rest-like disease in patients who develop this brain tumour.
Medulloblastoma
Medulloblastoma arises from the developing cerebellum and adjoining structures, and is the most common malignant brain tumour of childhood. It represents four distinct disease entities: the sonic hedgehog (SHH) and wingless-related integration site (WNT) subgroups are named based on the predominant signalling pathway driving tumorigenesis, whereas group 3 and group 4 medulloblastoma exhibit a more pleiotropic aetiology and do not have an easily identifiable oncogenic driver pathway (Figure 4).
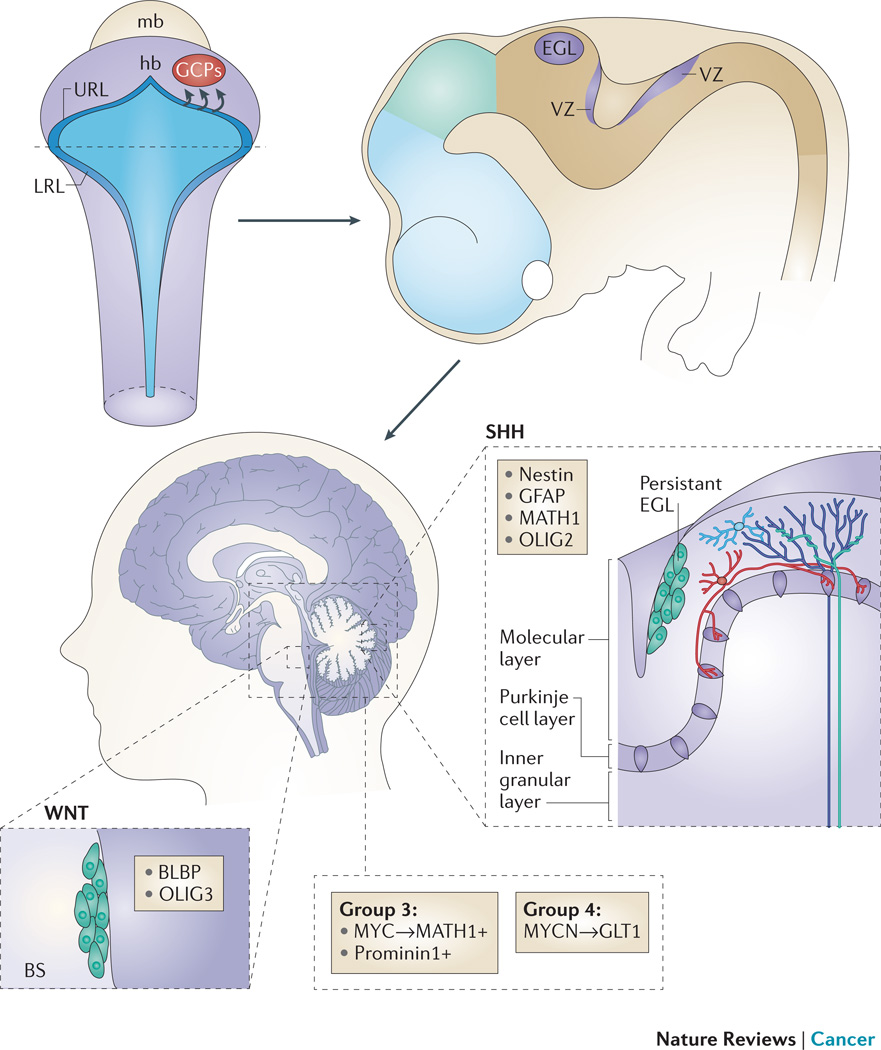
Figure 4. Cerebellar development and embryonal origin of medulloblastoma.
The upper (anterior) rhombic lip (URL) is a germinal zone of proliferating granule cell precursors (GCPs) that migrate rostrally to form the external granular layer (EGL). A second germinal centre is the ventricular zone (VZ), which gives rise to Purkinje cells and several other types of cerebellar interneurons. Sonic hedgehog (SHH) tumours may arise from persistent cells of the EGL that have not migrated to the internal granular layer (IGL), while Wnt tumours may originate from persistent cells of the ventricular zone in the brainstem (BS). Group 3 and 4 medulloblastomas likely arise from neural stem cells of the hindbrain.
Cerebellar development and medulloblastoma tumorigenesis
The fully developed cerebellum comprises an outer cell-sparse cortical layer (molecular layer), a Purkinje cell layer (PCL), and an internal granular layer (IGL), the latter is comprised of cerebellar granule cells (GCs) that form excitatory connections with Purkinje neurons95. Developmentally, the cerebellar anlage arises from the dorsal part of the anterior hindbrain, delineated by expression of the homeobox proteins orthodenticle homologue 2 (OTX2) anteriorly and HOXA2 posteriorly96, 97. OTX2 is a candidate driver for some group 3 medulloblastomas as it is amplified in 20% of these tumours, and is frequently overexpressed in SHH-independent subgroups98. Expression of OTX2 is abundant in the developing brain and silenced in the adult brain. OTX2 has recently been shown to repress differentiation in medulloblastoma cells, suggesting that amplification of OTX2 could generate a rest-like disease99. The observation that OTX2 also drives proliferation and can upregulate MYC, suggests that OTX2 might also be important in transforming rest-like cells into medulloblastoma98, 100.
The first germinal centre of the developing cerebellum initiates along the fourth ventricle in the dorsomedial ventricular zone (VZ), and gives rise to Purkinje cells and several other types of cerebellar interneurons101. Neural tube closure forms the rhombic lip, the anterior portion of which provides a second germinal zone of rapidly proliferating cells that express radial glial markers and gives rise to protein atonal homologue 1 (or MATH1)-positive granule cell progenitors (GCPs)102, 103. From 24 to 40 weeks gestation in humans, the volume of the developing cerebellum enlarges 5-fold, corresponding to more than a 30-fold increase in cerebellar cortex surface area104, 105. This expansion is largely imparted by the rapid proliferation of GCPs that give rise to cerebellar GCs and almost all cerebellar cortical neurons101, 106. Proliferation of these neuronal progenitors depends on SHH produced by Purkinje neurons: inhibiting SHH signalling blocks the proliferation and migration of GCPs107. GCPs migrate rostrally along the surface of the developing cerebellum to form the external granule layer (EGL), where they continue to divide and eventually migrate inward to form the IGL, with consequent depletion of the EGL108, 109. This process of GCP maturation and concurrent spontaneous cell death of excess GCPs is complete by the postnatal age of 20 months in children110.
The connection between SHH signalling and medulloblastoma was initially made in the context of Gorlin syndrome, an autosomal dominant disorder that causes developmental defects and predisposes individuals to several cancers, including medulloblastoma111. Fine mapping revealed the putative gene to be highly homologous to the Drosophila gene Patched (PTCH1)112. Loss of chromosome 9q, which contains PTCH1, occurs in ~30% of all SHH tumours113. The PTCH1 protein is an essential negative regulator of SHH signalling, thus mice heterozygous for Ptch1 develop cerebellar medulloblastoma114. Other germline mutations that predispose children to SHH medulloblastoma occur due to loss-of-function mutation in Suppressor of fused homolog (SUFU), a negative regulator of the SHH signalling115. Amplification of GLI family zinc finger 2 (GLI2), a positive regulator of the SHH signal, is also implicated in medulloblastoma development116. In a mouse model of SHH-induced medulloblastoma, tumour formation was preceded by GCP hyperplasia in the first week of life which regressed before later emerging as a malignant tumour117, in a manner similar to neuroblast rests and neuroblastoma in the Th-MYCN mice15. However, in vivo labelling studies following a rest cell to tumour transformation have not yet been performed in either model. In vitro, the murine GCPs were resistant to SHH withdrawal and this feature was partially MYCN-dependent117. Neural stem cells (NSCs) and GCPs have repeatedly been shown to be the cell of origin for SHH medulloblastoma118–122.
MYCN and its homologue MYC have crucial roles in all medulloblastoma subgroups. Brain-specific germline deletion of mouse Mycn results in cerebellar dysplasia123. Expression of MYCN is essential for SHH medulloblastoma in mouse models124 and amplification of MYCN marks a subset of poor-outcome SHH-driven human tumours125. Moreover, tumorigenic SHH signaling markedly increases MYCN expression in GCP cells due to effects on MYCN protein stability117. MYCN is also expressed in WNT and group 4 tumours, with targeted expression of MYCN or MYC driving SHH-independent medulloblastoma in transgenic mice126–128. Group 3 tumours express MYC and have amplification of MYC rather than MYCN129. Thus, there are many similarities between the role of MYCN in generating neuroblast rests and GCP hyperplasia in neuroblastoma and medulloblastoma, respectively.
WNT signalling has many roles in neural development and aberrant WNT signalling in NSCs of the cerebellar ventricular zone induces transient proliferation and impaired differentiation130. Individuals with Familiar Adenomatous Polyposis (FAP) and WNT pathway-driven colorectal cancer also have an increased risk of developing medulloblastoma131. Mutations that increase WNT signalling in members of the WNT signalling pathway have been described in sporadic medulloblastomas132–134. Recent data from the Medulloblastoma Advanced Genomics International Consortium showed that β-catenin (CTNNB1) displayed canonical exon 3 deletion in the vast majority (70–80%) of WNT-subgroup medulloblastoma116, 133, 135.
Insight into the cell of origin for WNT medulloblastomas comes from two mouse models, both involving the conditional expression of degradation-resistant β-catenin under the control of fatty acid binding protein 7 (Fabp7; also known as Blbp) in the context of Trp53 deletion (Blbp-cre:Ctnnb1:Trp53). The addition of MYC expression increased penetrance from 4% (Trp53+/-) and 15% (_Trp53_-/-) to 83%, suggesting that activation of β-catenin and WNT signalling alone are only weak oncogenic events136, 137. These MYC-driven medulloblastomas were evident in young mice and were formed from BLBP+, OLIG3+ mossy-fibre neuron precursors that are present in the brainstem between E11.5 and E15.5, consistent with the radiological observation that WNT medulloblastomas are frequently located within the fourth ventricle and infiltrate the dorsal surface of the brainstem126, 137, 138. This observation was consistent with the model of OLIG3+ neural precursors representing a brain stem rest disease preceding WNT medulloblastoma.
The pathogenesis of group 3 and group 4 medulloblastoma is less clear, yet they account for over 60% of all medulloblastoma cases139. Targeted inducible expression of MYCN to the postnatal cerebellum using the glial high affinity glutamate transporter (Glt1) promoter (Glt1-tTA:TRE-MYCN-Luc, or GTML mice) leads to tumours with a transcriptional profile of group 3 medulloblastoma126. GTML mice had a normal EGL, suggesting that group 3 medulloblastoma could originate from a cellular population that is distinct from SHH medulloblastoma. However, proliferating GCPs isolated from cyclin-dependent kinase inhibitor 2C (Cdkn2c)-/-, _Trp53_-/-, Math1-GFP mice generate group 3 medulloblastoma when transformed with MYC, but not MYCN127. Furthermore, group 3 tumours have been modelled through MYC transformation of postnatal cerebellar stem cells marked by the expression of prominin 1 (also known as CD133) and lack of neuronal or glial markers128. Thus, it appears that a single oncogene can give rise to multiple different medulloblastoma tumour subtypes, depending on the developmental age and anatomical origin of the transformed cell, indicating that susceptibility to medulloblastoma is highly dependent on embryonal site and stage140.
Unifying features of cancers with a prenatal origin
The work of Bishop and Varmus defined the forces driving human cancer as aberrations of our normal adult self 141, 142. Embryonal cancer initiation appears to be an aberration of our prenatal self. We propose here a new definition of rest cells which includes all cancers with a prenatal origin whether they arise directly from an embryonal cell, or a more mature prenatal cell which has acquired pathological properties that favour survival in the postnatal environment. Using this new definition of rests, neuroblastoma and TMD appears to initiate prenatally in cells which display resistance to cell death, numerical excess, and differentiation arrest at a stage of tissue ontogeny several steps distal to the earliest progenitor cell for each organ (Figure 5). We hypothesise that B-ALL cells might also pass through this phase during evolution toward the frankly malignant state. Mouse models suggest similar features exist for some medulloblastoma subtypes. Indeed, much of the support for rest-like disease in TMD, B-ALL and medulloblastoma, comes from mouse models. Thus, these findings raise interesting questions, but do not indicate definitively that the same pathways operate in humans. We propose that rests or rest-like cells are groups of abnormal embryonal cells which have undergone changes that allow them to persist through cell deletion signals in the postnatal environment. Yet they retain the capacity for self-destruction in the postnatal milieu unless other tumorigenic factors intervene to cause genomic instability, a feature which may offer an opportunity for rest deletion therapy. Neuroblastoma and TMD modelling indicates that all rest cells are not equal at birth, with only a small number undergoing transformation after later exposure to unidentified postnatal environmental influences15, 77. The properties that permit some rest cells to remain dormant after birth when other adjacent rest cells are dying is currently unknown.
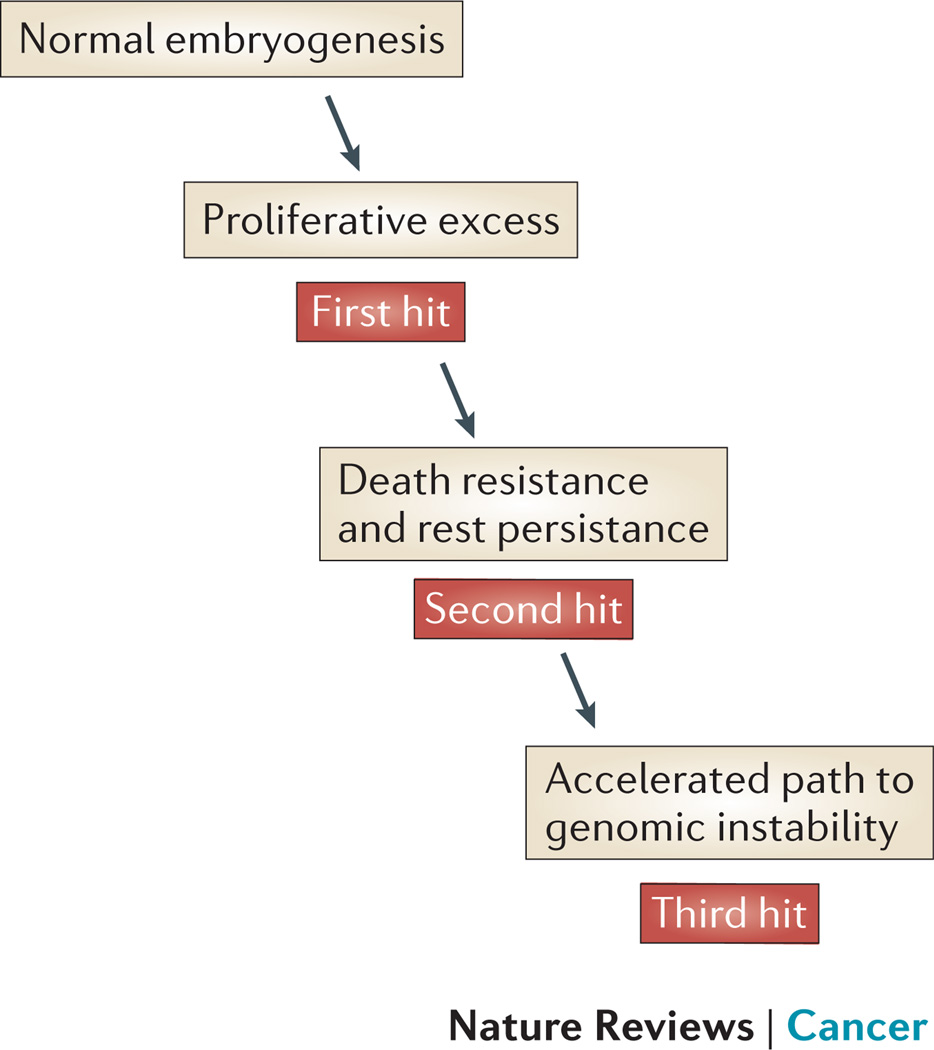
Figure 5. A model of embryonal tumorigenesis.
We propose that each of the four embryonal malignancies with evidence of a prenatal origin (neuroblastoma, ML-DS, B-ALL, medulloblastoma) share common features: (i) a prenatal proliferative excess in the tissue of origin; (ii) a cell intrinsic mechanism for surviving the hostile early postnatal environment; (iii) an accelerated pathway toward genomic instability.
For neuroblastoma and medulloblastoma, the timing and location of the rest-like disease in utero coincides with a point in the tissue-specific maturation pathway when rapid cell expansion is required, quickly followed by an orderly transition to a terminally differentiated state. Substantial cell death must occur during the final shaping of the nervous system. It is resistance to these death cues which must accompany an inappropriate replication stimulus, such as continued or high-level MYCN expression. We postulate that individual variation in factors required to: negatively regulate MYCN expression in specific tissues during embryogenesis, activate the apoptosis or senescence response to aberrant MYCN expression, or activate death responses imposed by trophic factor withdrawal, may explain individual susceptibility to rest disease.
From early in embryogenesis, haematopoietic precursors are in a continual state of replication and self-renewal, in contrast to terminally differentiated sympathetic ganglia and cerebellar neurons. Thus, while both TMD and B-ALL may not arise directly from an embryonal cell, the initial steps in each pathway toward postnatal malignancy involve progenitor cells acquiring characteristics that are favoured in the prenatal development. For TMD, and possibly B-ALL, the timing of rest initiation coincides with a unique predominance of foetal liver haematopoiesis. In the case of TMD, low interferon responses may be permissive for the survival of trisomy 21-primed and GATA1s mutant MKPs beyond the perinatal period in the non-permissive environment of the bone marrow 75.
The rest-to-cancer cell transition occurs in early childhood indicating that some rest cells must have the capacity for rapid progression toward genomic instability. One explanation for the speed of this process is inherited germline loss-of-function mutation in one allele of a tumour suppressor gene143. Another recently proposed mechanism is chromothripsis, whereby a single catastrophic event can lead to massive genomic rearrangement affecting more than one chromosome, rather than the incremental acquisition of single oncogenic mutations over decades144–146. An additional factor which may accelerate progression from rest to cancer are several recently described feed-forward loops, which increase the expression of MYCN to very high levels through the effects of SIRT1, SIRT2 and aurora kinase A on MYCN protein stability21, 22, 147. We propose that additional factors favouring rapid malignant transformation of rest cells are those properties of growth and self-renewal that are inherent in some embryonal cells.
Although chemotherapy, surgery and radiotherapy have improved the cure rates for childhood cancer, this has come at a significant cost in terms of the severe acute and long-term side-effects of therapy, and, considerable consumption of the paediatric health economy. Early evidence from screening trials in children at increased risk of cancer development due to germline p53 mutations has revived interest in strategies for child cancer prevention148, 149. While early detection of neuroblastoma using increased urine catecholamines failed to detect all neuroblastoma, one can envisage greater success when genomic susceptibility markers are incorporated into predictive algorithms for postnatal rest disease. Rest deletion therapy aimed at transiently restoring p53-mediated or interferon-mediated cell death responses or blocking MYCN, ALK, PI3K or MAPK signals in rest cells is an exciting strategy for childhood cancer prevention, which must first be demonstrated to be effective in mouse models of embryonal cancer. It is apparent that, with current models and a clearer understanding of the prenatal mis-steps that initiate rest disease, these novel strategies can be contemplated for the first time.
Acknowledgements
GMM, BBC, DC, and TL are supported by grants from the Australian NHMRC, Cancer Institute NSW, Cancer Council NSW, Steven Walter Children’s Cancer Foundation. TL is an ARC Research Fellow. MKM is supported by a fellowship from the Kids Cancer Project. JGM and WAW are supported by NIH grants F30CA174154, U01CA176287, R01CA133091, R01CA102321, R01CA148699, R01CA159859, U54CA163155, P01CA081403, Katie Dougherty, Pediatric Brain Tumor and Samuel G. Waxman Foundations. Sydney Children’s Hospital and Children’s Cancer Institute Australia for Medical Research are affiliated with the University of New South Wales. The authors apologize to the numerous colleagues whose important contributions could not be included in this Review owing to space limitations.
Glossary
Neural crest
is a transient collection of multipotent embryonic progenitors in the developing ectoderm that gives rise to a multitude of different cell types including melanocytes, craniofacial chondrocytes and osteocytes, smooth muscle myocytes and peripheral nervous system neurons
Somites
are bilaterally paired blocks of mesoderm that form along the anterior-posterior axis of the developing embryo
Anlage
the initial clustering of embryonic cells from which an organ, or part of an organ, will form
Rostrally
situated toward the oral or nasal region, or in the case of the brain, toward the tip of the frontal lobe
Thrombocytopaenia
a platelet number below the normal range, in the peripheral blood
Neonatal blood spots
cards with drops of blood collected from newborn baby’s heel, which are used for screening neonates for rare but serious metabolic conditions
Innate B-1 cells
B-1 cells with innate sensing and responding properties
Marginal zone B cells
B cells from the marginal zone of the spleen, a unique lymphoid area located at the interface between the circulation and the immune system
Adaptive B-2 B cells
B-2 cells responsible for the production of antibodies during adaptive immunity
Chromothripsis
process whereby a single catastrophic event within the genome leads to multiple genetic alterations across one or more chromosomes
Footnotes
Competing interest’s statement: The authors declare no conflicts or competing interests.
References
- 1.Valent P, et al. Cancer stem cell definitions and terminology: the devil is in the details. Nat Rev Cancer. 2012;12:767–775. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 2.Gruhn B, et al. Prenatal origin of childhood acute lymphoblastic leukemia, association with birth weight and hyperdiploidy. Leukemia. 2008;22:1692–1697. doi: 10.1038/leu.2008.152. [DOI] [PubMed] [Google Scholar]
- 3.Pine SR, et al. Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood. 2007;110:2128–2131. doi: 10.1182/blood-2007-01-069542. [DOI] [PubMed] [Google Scholar]
- 4.Nuchtern JG, et al. A prospective study of expectant observation as primary therapy for neuroblastoma in young infants: a Children's Oncology Group study. Annals of Surgery. 2012;256:573–580. doi: 10.1097/SLA.0b013e31826cbbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckwith JB. Precursor lesions of Wilms tumor: clinical and biological implications. Med Pediatr Oncol. 1993;21:158–168. doi: 10.1002/mpo.2950210303. [DOI] [PubMed] [Google Scholar]
- 6.Cohnheim J. Congenitales, quergestreiftes Muskelsarkom der Nieren. Virchows Archiv. 1875;65:64–69. [Google Scholar]
- 7.Durante F. Nesso fisio-patologico tra la struttura dei nei materni e la genesi di alcuni tumori maligni. Arch Memo Observ Chir Prat. 1874;11:217–226. [Google Scholar]
- 8.Virchow R. Die multiloculäre, ulcerirende Echinokokkengeschwulst der Leber. Verhandlungen der Physicalisch-Medicinischen Gesellschaft. 1855:84–95. [Google Scholar]
- 9.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 10.Eaton KW, Tooke LS, Wainwright LM, Judkins AR, Biegel JA. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr Blood Cancer. 2011;56:7–15. doi: 10.1002/pbc.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nature Reviews. Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 12.Maris JM. Recent advances in neuroblastoma. New England Journal of Medicine. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber K. The sympathoadrenal cell lineage: specification, diversification, and new perspectives. Dev Biol. 2006;298:335–343. doi: 10.1016/j.ydbio.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman KA, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 15.Hansford LM, et al. Mechanisms of embryonal tumor initiation: distinct roles for MycN expression and MYCN amplification. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12664–12669. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wartiovaara K, Barnabe-Heider F, Miller FD, Kaplan DR. N-myc promotes survival and induces S-phase entry of postmitotic sympathetic neurons. J Neurosci. 2002;22:815–824. doi: 10.1523/JNEUROSCI.22-03-00815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 18.Zhu S, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362–373. doi: 10.1016/j.ccr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calao M, et al. Direct effects of Bmi1 on p53 protein stability inactivates oncoprotein stress responses in embryonal cancer precursor cells at tumor initiation. Oncogene. 2013;32:3616–3626. doi: 10.1038/onc.2012.368. [DOI] [PubMed] [Google Scholar]
- 20.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO Journal. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall GM, et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genetics. 2011;7:e1002135. doi: 10.1371/journal.pgen.1002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu PY, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death and Differentiation. 2013;20:503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy DJ, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berry T, et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell. 2012;22:117–130. doi: 10.1016/j.ccr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reissmann E, et al. Involvement of bone morphogenetic protein-4 and bone morphogenetic protein-7 in the differentiation of the adrenergic phenotype in developing sympathetic neurons. Development. 1996;122:2079–2088. doi: 10.1242/dev.122.7.2079. [DOI] [PubMed] [Google Scholar]
- 26.Schneider C, Wicht H, Enderich J, Wegner M, Rohrer H. Bone morphogenetic proteins are required in vivo for the generation of sympathetic neurons. Neuron. 1999;24:861–870. doi: 10.1016/s0896-6273(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 27.Mosse YP, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75:727–730. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trochet D, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. American Journal of Human Genetics. 2004;74:761–764. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourdeaut F, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Cancer Lett. 2005;228:51–58. doi: 10.1016/j.canlet.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 30.Reiff T, et al. Neuroblastoma phox2b variants stimulate proliferation and dedifferentiation of immature sympathetic neurons. J Neurosci. 2010;30:905–915. doi: 10.1523/JNEUROSCI.5368-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam G, et al. MYCN promotes the expansion of Phox2B-positive neuronal progenitors to drive neuroblastoma development. Am J Pathol. 2009;175:856–866. doi: 10.2353/ajpath.2009.090019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stemcell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 33.Molenaar JJ, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 35.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 36.Murray MJ, et al. LIN28 Expression in malignant germ cell tumors downregulates let-7 and increases oncogene levels. Cancer Res. 2013;73:4872–4884. doi: 10.1158/0008-5472.CAN-12-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiff T, et al. Midkine and Alk signaling in sympathetic neuron proliferation and neuroblastoma predisposition. Development. 2011;138:4699–4708. doi: 10.1242/dev.072157. [DOI] [PubMed] [Google Scholar]
- 38.Cheng LY, et al. Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell. 2011;146:435–447. doi: 10.1016/j.cell.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 39.Mosse YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janoueix-Lerosey I, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967-U951. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 41.Chen YY, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971-U956. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 42.De Brouwer S, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res. 2010;16:4353–4362. doi: 10.1158/1078-0432.CCR-09-2660. [DOI] [PubMed] [Google Scholar]
- 43.Heukamp LC, et al. Targeted Expression of Mutated ALK Induces Neuroblastoma in Transgenic Mice. Science Translational Medicine. 2012;4:ra91. doi: 10.1126/scitranslmed.3003967. [DOI] [PubMed] [Google Scholar]
- 44.Schulte JH, et al. MYCN and ALKF1174L are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene. 2012;21:1059–1065. doi: 10.1038/onc.2012.106. [DOI] [PubMed] [Google Scholar]
- 45.De Preter K, et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biology. 2006;7:17. doi: 10.1186/gb-2006-7-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beckwith JB, Perrin EV. In Situ Neuroblastomas: A Contribution to the Natural History of Neural Crest Tumors. Am J Pathol. 1963;43:1089–1104. [PMC free article] [PubMed] [Google Scholar]
- 47.Woods WG, et al. A population-based study of the usefulness of screening for neuroblastoma. Lancet. 1996;348:1682–1687. doi: 10.1016/S0140-6736(96)06020-5. [DOI] [PubMed] [Google Scholar]
- 48.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 49.Roy A, Roberts I, Norton A, Vyas P. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: a multi-step model of myeloid leukaemogenesis. British Journal of Haematology. 2009;147:3–12. doi: 10.1111/j.1365-2141.2009.07789.x. [DOI] [PubMed] [Google Scholar]
- 50.Brodeur GM, Dahl GV, Williams DL, Tipton RE, Kalwinsky DK. Transient leukemoid reaction and trisomy 21 mosaicism in a phenotypically normal newborn. Blood. 1980;55:691–693. [PubMed] [Google Scholar]
- 51.Hasle H, et al. A pediatric approach to the WHO classification of myelodysplastic and myeloproliferative diseases. Leukemia. 2003;17:277–282. doi: 10.1038/sj.leu.2402765. [DOI] [PubMed] [Google Scholar]
- 52.Massey GV, et al. A prospective study of the natural history of transient leukemia (TL) in neonates with Down syndrome (DS): Children's Oncology Group (COG) study POG-9481. Blood. 2006;107:4606–4613. doi: 10.1182/blood-2005-06-2448. [DOI] [PubMed] [Google Scholar]
- 53.Klusmann JH, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008;111:2991–2998. doi: 10.1182/blood-2007-10-118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tunstall-Pedoe O, et al. Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations. Blood. 2008;112:4507–4511. doi: 10.1182/blood-2008-04-152967. [DOI] [PubMed] [Google Scholar]
- 55.Polski JM, et al. Acute megakaryoblastic leukemia after transient myeloproliferative disorder with clonal karyotype evolution in a phenotypically normal neonate. Journal of Pediatric Hematology/Oncology. 2002;24:50–54. doi: 10.1097/00043426-200201000-00014. [DOI] [PubMed] [Google Scholar]
- 56.Kirsammer G, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood. 2008;111:767–775. doi: 10.1182/blood-2007-04-085670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maclean GA, et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17567–17572. doi: 10.1073/pnas.1215468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chou ST, et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17573–17578. doi: 10.1073/pnas.1211175109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dore LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118:231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bourquin JP, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113:3337–3347. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salek-Ardakani S, et al. ERG is a megakaryocytic oncogene. Cancer Research. 2009;69:4665–4673. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 63.Birger Y, et al. Perturbation of fetal hematopoiesis in a mouse model of Down syndrome's transient myeloproliferative disorder. Blood. 2013;122:988–998. doi: 10.1182/blood-2012-10-460998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elagib KE, et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 65.Gurbuxani S, Vyas P, Crispino JD. Recent insights into the mechanisms of myeloid leukemogenesis in Down syndrome. Blood. 2004;103:399–406. doi: 10.1182/blood-2003-05-1556. [DOI] [PubMed] [Google Scholar]
- 66.Taub JW, et al. Prenatal origin of GATA1 mutations may be an initiating step in the development of megakaryocytic leukemia in Down syndrome. Blood. 2004;104:1588–1589. doi: 10.1182/blood-2004-04-1563. [DOI] [PubMed] [Google Scholar]
- 67.Li Z, et al. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nature Genetics. 2005;37:613–619. doi: 10.1038/ng1566. [DOI] [PubMed] [Google Scholar]
- 68.Hitzler JK, Cheung J, Li Y, Scherer SW, Zipursky A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood. 2003;101:4301–4304. doi: 10.1182/blood-2003-01-0013. [DOI] [PubMed] [Google Scholar]
- 69.Rainis L, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood. 2003;102:981–986. doi: 10.1182/blood-2002-11-3599. [DOI] [PubMed] [Google Scholar]
- 70.Alford KA, et al. Analysis of GATA1 mutations in Down syndrome transient myeloproliferative disorder and myeloid leukemia. Blood. 2011;118:2222–2238. doi: 10.1182/blood-2011-03-342774. [DOI] [PubMed] [Google Scholar]
- 71.Kanezaki R, et al. Down syndrome and GATA1 mutations in transient abnormal myeloproliferative disorder: mutation classes correlate with progression to myeloid leukemia. Blood. 2010;116:4631–4638. doi: 10.1182/blood-2010-05-282426. [DOI] [PubMed] [Google Scholar]
- 72.Zipursky A. Transient leukaemia--a benign form of leukaemia in newborn infants with trisomy 21. British Journal of Haematology. 2003;120:930–938. doi: 10.1046/j.1365-2141.2003.04229.x. [DOI] [PubMed] [Google Scholar]
- 73.Muramatsu H, et al. Risk factors for early death in neonates with Down syndrome and transient leukaemia. British Journal of Haematology. 2008;142:610–615. doi: 10.1111/j.1365-2141.2008.07231.x. [DOI] [PubMed] [Google Scholar]
- 74.Roy A, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17579–17584. doi: 10.1073/pnas.1211405109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woo AJ, et al. Developmental differences in IFN signaling affect GATA1s-induced megakaryocyte hyperproliferation. J Clin Invest. 2013 doi: 10.1172/JCI40609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nikolaev SI, et al. Exome sequencing identifies putative drivers of progression of transient myeloproliferative disorder to AMKL in infants with Down syndrome. Blood. 2013;122:554–561. doi: 10.1182/blood-2013-03-491936. [DOI] [PubMed] [Google Scholar]
- 77.Saida S, et al. Clonal selection in xenografted TAM recapitulates the evolutionary process of myeloid leukemia in Down syndrome. Blood. 2013;121:4377–4387. doi: 10.1182/blood-2012-12-474387. [DOI] [PubMed] [Google Scholar]
- 78.Roberts I, et al. GATA1-mutant clones are frequent and often unsuspected in babies with Down syndrome: identification of a population at risk of leukemia. Blood. 2013 doi: 10.1182/blood-2013-07-515148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ford AM, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 80.Wiemels JL, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 81.Teuffel O, et al. Prenatal origin of separate evolution of leukemia in identical twins. Leukemia. 2004;18:1624–1629. doi: 10.1038/sj.leu.2403462. [DOI] [PubMed] [Google Scholar]
- 82.Taub JW, et al. High frequency of leukemic clones in newborn screening blood samples of children with Bprecursor acute lymphoblastic leukemia. Blood. 2002;99:2992–2996. doi: 10.1182/blood.v99.8.2992. [DOI] [PubMed] [Google Scholar]
- 83.Eguchi-Ishimae M, et al. Breakage and fusion of the TEL (ETV6) gene in immature B lymphocytes induced by apoptogenic signals. Blood. 2001;97:737–743. doi: 10.1182/blood.v97.3.737. [DOI] [PubMed] [Google Scholar]
- 84.McHale CM, et al. Prenatal origin of TEL-AML1-positive acute lymphoblastic leukemia in children born in California. Genes Chromosomes Cancer. 2003;37:36–43. doi: 10.1002/gcc.10199. [DOI] [PubMed] [Google Scholar]
- 85.Olsen M, et al. Preleukemic TEL-AML1-positive clones at cell level of 10(-3) to 10(-4) do not persist into adulthood. Journal of Pediatric Hematology/Oncology. 2006;28:734–740. doi: 10.1097/01.mph.0000243652.33561.0f. [DOI] [PubMed] [Google Scholar]
- 86.Hong D, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 87.Tuszuki S, Seto M. TEL (ETV6)-AML1 (RUNX1) initiates self-renewing fetal pro-B cells in association with a transcriptional program shared with embryonic stem cells in mice. Stem Cells. 2013;31:236–247. doi: 10.1002/stem.1277. [DOI] [PubMed] [Google Scholar]
- 88.Torrano V, Procter J, Cardus P, Greaves M, Ford AM. ETV6-RUNX1 promotes survival of early B lineage progenitor cells via a dysregulated erythropoietin receptor. Blood. 2011;118:4910–4918. doi: 10.1182/blood-2011-05-354266. [DOI] [PubMed] [Google Scholar]
- 89.van der Weyden L, et al. Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice. Blood. 2011;118:1041–1051. doi: 10.1182/blood-2011-02-338848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ford AM, et al. The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells. Journal of Clinical Investigation. 2009;119:826–836. doi: 10.1172/JCI36428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cazzaniga G, et al. Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood. 2011;118:5559–5564. doi: 10.1182/blood-2011-07-366542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chuk MK, McIntyre E, Small D, Brown P. Discordance of MLL-rearranged (MLL-R) infant acute lymphoblastic leukemia in monozygotic twins with spontaneous clearance of preleukemic clone in unaffected twin. Blood. 2009;113:6691–6694. doi: 10.1182/blood-2009-01-202259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Powell BC, et al. Identification of TP53 as an acute lymphocytic leukemia susceptibility gene through exome sequencing. Pediatr Blood Cancer. 2013;60:E1–E3. doi: 10.1002/pbc.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ito M. Control of mental activities by internal models in the cerebellum. Nature Reviews Neuroscience. 2008;9:304–313. doi: 10.1038/nrn2332. [DOI] [PubMed] [Google Scholar]
- 96.Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing rostral brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- 97.Wingate RJ, Hatten ME. The role of the rhombic lip in avian cerebellum development. Development. 1999;126:4395–4404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- 98.Adamson DC, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Research. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bai RY, Staedtke V, Lidov HG, Eberhart CG, Riggins GJ. OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells. Cancer Research. 2012;72:5988–6001. doi: 10.1158/0008-5472.CAN-12-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bunt J, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. International Journal of Cancer. 2012;131:E21–E32. doi: 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- 101.Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annual Review of Neuroscience. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 102.Morales D, Hatten ME. Molecular markers of neuronal progenitors in the embryonic cerebellar anlage. Journal of Neuroscience. 2006;26:12226–12236. doi: 10.1523/JNEUROSCI.3493-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Developmental Biology. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang CH, Chang FM, Yu CH, Ko HC, Chen HY. Assessment of fetal cerebellar volume using threedimensional ultrasound. Ultrasound in Medicine & Biology. 2000;26:981–988. doi: 10.1016/s0301-5629(00)00225-8. [DOI] [PubMed] [Google Scholar]
- 105.Volpe JJ. Cerebellum of the premature infant: rapidly developing, vulnerable, clinically important. Journal of Child Neurology. 2009;24:1085–1104. doi: 10.1177/0883073809338067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Andersen BB, Korbo L, Pakkenberg B. A quantitative study of the human cerebellum with unbiased stereological techniques. Journal of Comparative Neurology. 1992;326:549–560. doi: 10.1002/cne.903260405. [DOI] [PubMed] [Google Scholar]
- 107.Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 108.Lee A, et al. Isolation of neural stem cells from the postnatal cerebellum. Nature Neuroscience. 2005;8:723–729. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 110.Raaf J, Kernohan JW. A study of the external granular layer in the cerebellum. The disappearance of the external granular layer and the growth of the molecular and internal granular layers in the cerebellum. Am. J. Anat. 1944;75:151–172. [Google Scholar]
- 111.Gailani MR, et al. Developmental defects in Gorlin syndrome related to a putative tumor suppressor gene on chromosome 9. Cell. 1992;69:111–117. doi: 10.1016/0092-8674(92)90122-s. [DOI] [PubMed] [Google Scholar]
- 112.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 113.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. Journal of Clinical Oncology. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 115.Brugieres L, et al. Incomplete penetrance of the predisposition to medulloblastoma associated with germline SUFU mutations. Journal of Medical Genetics. 2010;47:142–144. doi: 10.1136/jmg.2009.067751. [DOI] [PubMed] [Google Scholar]
- 116.Northcott PA, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thomas WD, et al. Patched1 deletion increases N-Myc protein stability as a mechanism of medulloblastoma initiation and progression. Oncogene. 2009;28:1605–1615. doi: 10.1038/onc.2009.3. [DOI] [PubMed] [Google Scholar]
- 118.Hallahan AR, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Research. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 119.Mao J, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Research. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Weiner HL, et al. Induction of medulloblastomas in mice by sonic hedgehog, independent of Gli1. Cancer Research. 2002;62:6385–6389. [PubMed] [Google Scholar]
- 121.Schuller U, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Browd SR, et al. N-myc can substitute for insulin-like growth factor signaling in a mouse model of sonic hedgehog-induced medulloblastoma. Cancer Research. 2006;66:2666–2672. doi: 10.1158/0008-5472.CAN-05-2198. [DOI] [PubMed] [Google Scholar]
- 125.Korshunov A, et al. Biological and clinical heterogeneity of MYCN-amplified medulloblastoma. Acta Neuropathologica. 2012;123:515–527. doi: 10.1007/s00401-011-0918-8. [DOI] [PubMed] [Google Scholar]
- 126.Swartling FJ, et al. Pleiotropic role for MYCN in medulloblastoma. Genes & Development. 2010;24:1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kawauchi D, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pei Y, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21:155–167. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kool M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathologica. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pei Y, et al. WNT signaling increases proliferation and impairs differentiation of stem cells in the developing cerebellum. Development. 2012;139:1724–1733. doi: 10.1242/dev.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kropilak M, Jagelman DG, Fazio VW, Lavery IL, McGannon E. Brain tumors in familial adenomatous polyposis. Diseases of the Colon & Rectum. 1989;32:778–782. doi: 10.1007/BF02562128. [DOI] [PubMed] [Google Scholar]
- 132.Taylor MD, et al. Failure of a medulloblastoma-derived mutant of SUFU to suppress WNT signaling. Oncogene. 2004;23:4577–4583. doi: 10.1038/sj.onc.1207605. [DOI] [PubMed] [Google Scholar]
- 133.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barker N, et al. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO Journal. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Momota H, Shih AH, Edgar MA, Holland EC. c-Myc and beta-catenin cooperate with loss of p53 to generate multiple members of the primitive neuroectodermal tumor family in mice. Oncogene. 2008;27:4392–4401. doi: 10.1038/onc.2008.81. [DOI] [PubMed] [Google Scholar]
- 137.Gibson P, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takebayashi H, et al. Non-overlapping expression of Olig3 and Olig2 in the embryonic neural tube. Mechanisms of Development. 2002;113:169–174. doi: 10.1016/s0925-4773(02)00021-7. [DOI] [PubMed] [Google Scholar]
- 139.Taylor MD, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathologica. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Swartling FJ, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to NMYC. Cancer Cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 142.Siddhartha M. The Emperor of All Maladies: A Biography of Cancer. SCRIBNER; New York: 2010. [Google Scholar]
- 143.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nature Reviews. Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 146.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 147.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 148.Masciari S, et al. F18-fluorodeoxyglucose-positron emission tomography/computed tomography screening in Li-Fraumeni syndrome. JAMA. 2008;299:1315–1319. doi: 10.1001/jama.299.11.1315. [DOI] [PubMed] [Google Scholar]
- 149.Custodio G, et al. Impact of neonatal screening and surveillance for the TP53 R337H mutation on early detection of childhood adrenocortical tumors. J Clin Oncol. 2013;31:2619–2626. doi: 10.1200/JCO.2012.46.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yoshimoto M, et al. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1468–1473. doi: 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hardy RR. B-1 B cell development. Journal of Immunology. 2006;177:2749–2754. doi: 10.4049/jimmunol.177.5.2749. [DOI] [PubMed] [Google Scholar]
- 152.Montecino-Rodriguez E, Dorshkind K. Formation of B-1 B cells from neonatal B-1 transitional cells exhibits NF-kB redundancy. Journal of Immunology. 2011;187:5712–5719. doi: 10.4049/jimmunol.1102416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 154.Li YS, Wasserman R, Hayakawa K, Hardy RR. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 155.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19-early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sitnicka E, et al. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J Exp Med. 2003;198:1495–1506. doi: 10.1084/jem.20031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sitnicka E, et al. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–472. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 158.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 159.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32:720–730. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 160.Wang JH, et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 161.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 163.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 164.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 165.Shah S, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bain G, et al. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 167.Borghesi L, et al. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Welinder E, et al. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci U S A. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J Immunol. 2008;181:383–392. doi: 10.4049/jimmunol.181.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1-Cbfbeta facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med. 2012;209:1255–1262. doi: 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Growney JD, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tsubono Y, Hisamichi S. A halt to neuroblastoma screening in Japan. New England Journal of Medicine. 2004;350:2010–2011. doi: 10.1056/NEJM200405063501922. [DOI] [PubMed] [Google Scholar]
- 173.Breslow N, Olshan A, Beckwith JB, Green DM. Epidemiology of Wilms tumor. Med Pediatr Oncol. 1993;21:172–181. doi: 10.1002/mpo.2950210305. [DOI] [PubMed] [Google Scholar]
- 174.Coppes MJ, Haber DA, Grundy PE. Genetic events in the development of Wilms' tumor. N Engl J Med. 1994;331:586–590. doi: 10.1056/NEJM199409013310906. [DOI] [PubMed] [Google Scholar]
- 175.Park S, et al. Inactivation of WT1 in nephrogenic rests, genetic precursors to Wilms' tumour. Nat Genet. 1993;5:363–367. doi: 10.1038/ng1293-363. [DOI] [PubMed] [Google Scholar]
- 176.Chen D, et al. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 177.Zhang J, Schweers B, Dyer MA. The first knockout mouse model of retinoblastoma. Cell Cycle. 2004;3:952–959. [PubMed] [Google Scholar]
- 178.Federico S, Brennan R, Dyer MA. Childhood cancer and developmental biology a crucial partnership. Curr Top Dev Biol. 2011;94:1–13. doi: 10.1016/B978-0-12-380916-2.00001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Abramson DH, et al. Rapid growth of retinoblastoma in a premature twin. Arch Ophthalmol. 2002;120:1232–1233. doi: 10.1001/archopht.120.9.1232. [DOI] [PubMed] [Google Scholar]
- 180.Lohmann DR, Gallie BL. Retinoblastoma: revisiting the model prototype of inherited cancer. Am J Med Genet C Semin Med Genet. 2004;129C:23–28. doi: 10.1002/ajmg.c.30024. [DOI] [PubMed] [Google Scholar]
- 181.Looijenga LH, Oosterhuis JW. Pathogenesis of testicular germ cell tumours. Rev Reprod. 1999;4:90–100. doi: 10.1530/ror.0.0040090. [DOI] [PubMed] [Google Scholar]
- 182.Bussey KJ, et al. Chromosome abnormalities of eighty-one pediatric germ cell tumors: sex-, age-, site-, and histopathology-related differences--a Children's Cancer Group study. Genes Chromosomes Cancer. 1999;25:134–146. [PubMed] [Google Scholar]
- 183.Isaacs H., Jr Perinatal (fetal and neonatal) germ cell tumors. J Pediatr Surg. 2004;39:1003–1013. doi: 10.1016/j.jpedsurg.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 184.Schindler JW, et al. TEL-AML1 corrupts hematopoietic stem cells to persist in the bone marrow and initiate leukemia. Cell Stem Cell. 2009;5:43–53. doi: 10.1016/j.stem.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 185.Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 187.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 188.Sevenet N, et al. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]